Research
OVERVIEW
Bacterial cells were long thought to be “bags of enzymes” with little or no intracellular organization. However, over the last three decades, the advent of fluorescent proteins and development of new imaging modalities (e.g. electron cryotomography, super-resolution light microscopy etc.) have revolutionized our understanding of the bacterial domain of life. To survive efficiently in its environment, the bacterial cell requires a high spatiotemporal organization both structurally and metabolically. They utilize sophisticated sensory and regulation pathways to orchestrate motility and cell-cell communication, assemble complex molecular machines to inject effectors into neighbouring bacterial/eukaryotic cells, amass nutrients inside specialized compartments, possess eukaryotic homologues of actin, tubulin and intermediate filaments, and even some of them own membrane bound organelles and distinct cellular compartmentalization. This paradigm shift in our understanding has opened up new opportunities to explore ever increasing internal complexities within bacterial cells!
Electron cryotomography (cryo-ET) has unrivalled power to visualize the native structure of macromolecular complexes in their mise-en-scène, i.e., inside ‘intact’ cells and thus provides unprecedented mechanistic and structural insights. In this method, plunge-frozen vitreous samples are iteratively imaged in a transmission electron microscope (TEM) as they are tilted, resulting in a stack of 2D images (tilt-series) that can be reconstructed into a 3D volume or a “tomogram”. With resolutions of ~2-5 nm, cryo-ET uniquely bridges the “resolution gap” between atomic structures and gross cellular morphology. Additionally, recent improvements in software, hardware and the implementation of improved subvolume averaging methods have allowed cryo-ET to enter the realm of subnanometer resolution. In addition to structural information, cryo-ET also provides pivotal insights into the distribution pattern of protein complexes in cells, their relative orientations and structural dynamics making this an ideal method for ‘visual proteomics’ and in-cell structural biology. We are harnessing this unique power of cryo-ET and combining it with correlative light and electron microscopy (CLEM), and Focused Ion Beam (FIB) milling to elucidate the structure and function of large bacterial macromolecular assemblies in their native context inside ‘living’ cells.
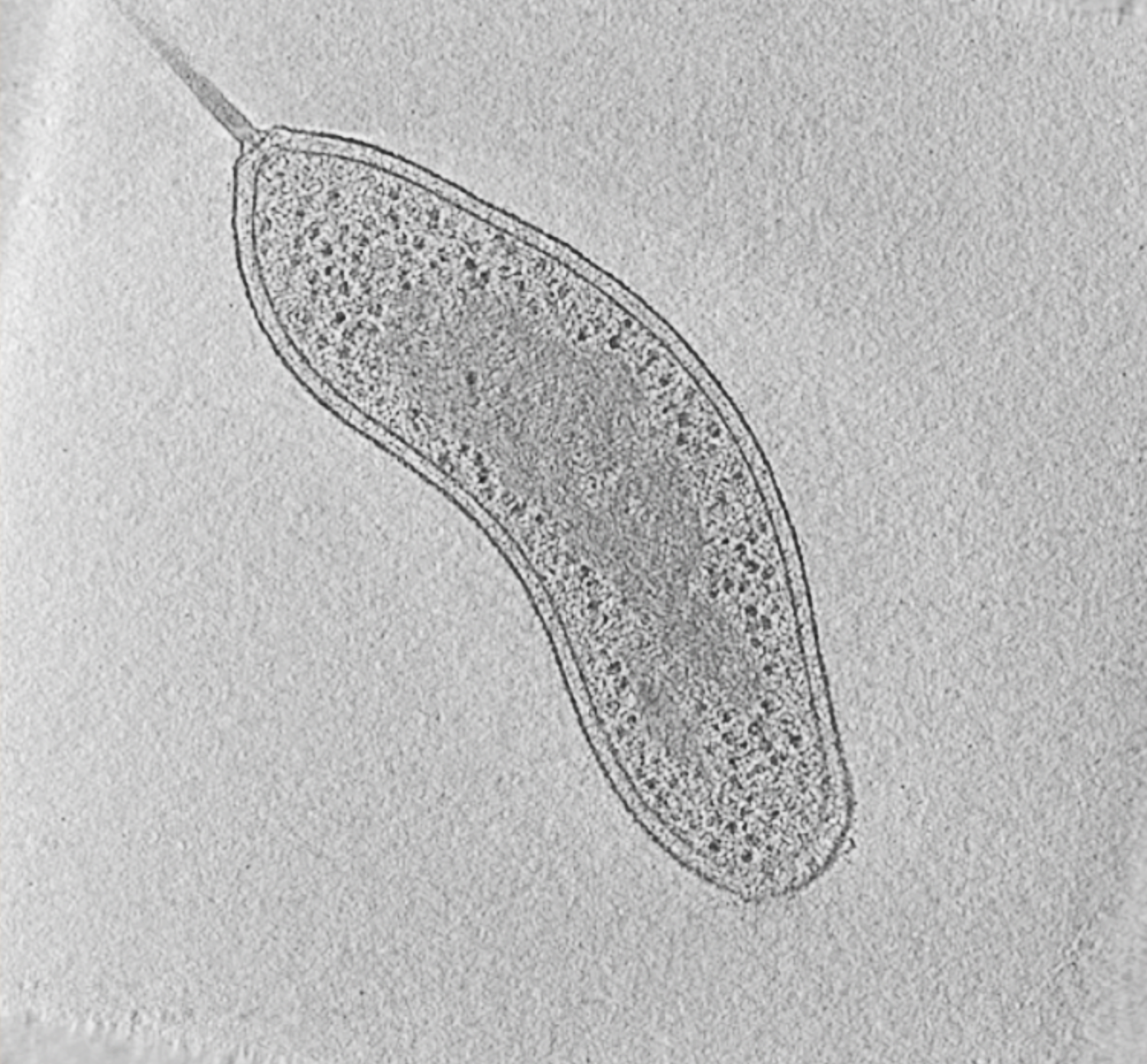
Fig. 1: Cryotomogram of an intact Bdellovibrio bacteriovorus cell. Visible cellular structures are segmented and annotated. Left: tomographic slice, right: segmented tomogram. Movie: courtesy of Grant Jensen lab.
We complement cryo-ET with other biochemical and biophysical techniques such as in vitro biochemical reconstitution, genetic and functional analysis, fluorescence imaging and high-resolution structural techniques such as X-ray crystallography and cryo-EM.
RESEARCH PROJECTS
We are particularly interested in bacterial molecular machines that are responsible for toxin delivery (bacterial secretion systems), and bacterial cytoskeletal (rather, bacterial ‘cytomotive’) filaments that execute a diverse array of cellular functions.

Bacterial Secretion systems:
Bacteria harbour at least nine different types of secretion systems to transfer macromolecules across cellular envelope. These are sophisticated multi-protein nanomachines that secrete myriads of substrates including proteins, nucleoprotein complexes and variety of small molecules and are central to pathogenesis of multiple human diseases. For example, many pathogenic bacteria utilize the Type III Secretion System (T3SS) to cause diseases such as typhoid (Salmonella), plague (Yersinia), dysentery (Shigella) etc. Other human pathogens employ the Type IV Secretion System (T4SS) to mediate gastric cancer (Helicobacter), brucellosis (Brucella), typhus and spotted fevers (Rickettsia), as well as Legionnaires’ disease (Legionella). The T4SS is also associated with the spread of antibiotic resistance, which currently presents a major threat to public health. Therefore, these molecular machines are attractive targets for drug developments to enrich our present repertoire of antibiotics. However, structural studies on these molecular machines are extremely challenging due to their large number of components, flexibility and tight integration into the bacterial cell envelope.
Recently, we used cryo-ET to decipher the in situ structure and molecular organization of the intact Legionella T4SS, responsible for Legionnaires’ disease. Our work revealed that the T4SS has an envelope spanning channel and a large periplasmic ‘secretion chamber’.
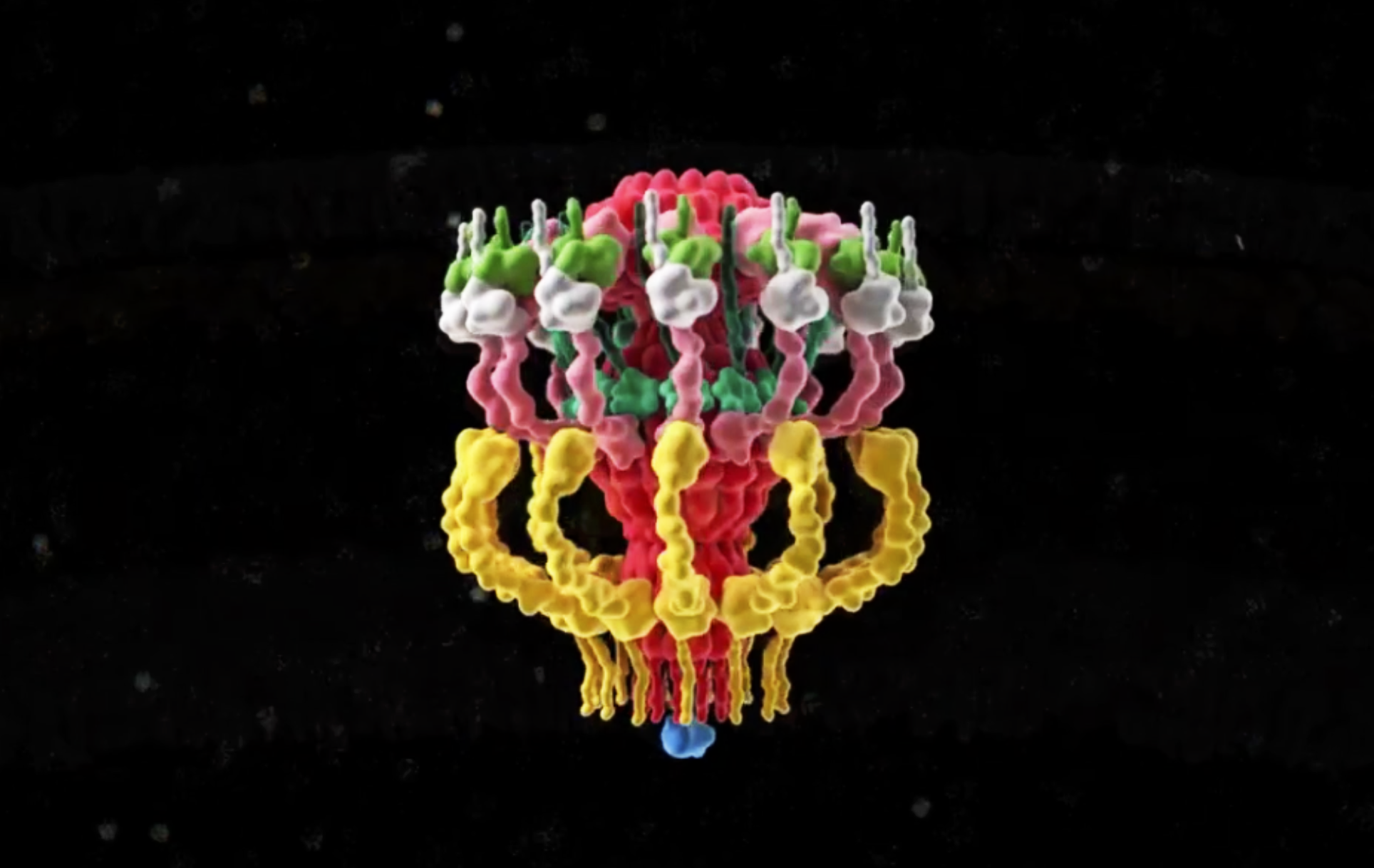
Fig. 2: 3D representation of the Dot/Icm complex showing a windowed secretion chamber (salmon, DotH; grey, DotD; green, DotK; and cyan, DotC), wings (yellow, DotF), a secretion channel (red, DotG) and the top-view of the complex. Cytoplasmic components are not shown. (Ghosal et al, Nature Microbiology).
Using a combination of genetic manipulation, immunofluorescence imaging and biochemical analysis, we also revealed the mechanism of polar targeting and biogenesis of this complex. This now sets the stage for investigating how this remarkable molecular machine is capable of secreting more than 300 different types of effectors and dissect its recondite modus operandi in the future.

Structure and function of bacterial ‘cytomotive’ filaments
Until early 1990s, cytoskeletal proteins were believed to be the hallmarks of eukaryotic cells. However, in the last three decades, the discovery of bacterial homologs of eukaryotic actin (e.g. MreB), tubulin (FtsZ), and intermediate filament (IF) (e.g. crescentin) proteins have dramatically changed our perception. Recent studies have revealed that the catalogue of bacterial cytoskeletal proteins is potentially much longer than previously thought. Bioinformatic analyses of published genomic data have shown that there are at least 35 distinct families of bacterial actins. Similarly, additional homologs of tubulin and IF have been also discovered. Growing evidence suggests that bacteria also harbour additional cytoskeletal filament systems with no eukaryotic homologs such as the Walker A Cytoskeletal ATPases (WACAs), bactofilins, and metabolic enzyme CTP Synthase filaments. In the ‘absence’ of motor proteins (e.g. kinesins and dyneins), these filamentous systems perform bacterial cell division, generate force, control cell shape, partition DNA and organize intracellular space. One of the key emerging difference between the bacterial and the eukaryotic cytoskeletal systems is that each of the bacterial filaments seem to perform one dedicated function while eukaryotic ones, by virtue of their interaction with a repertoire of adapters and regulatory proteins, perform numerous tasks. How and when this transition happened in evolution is still unclear. We need to investigate many more bacterial filament systems and their interactome to decipher how prokaryotic cytoskeleton was customised for multifunctionality during the evolution of complex eukaryotic cells.
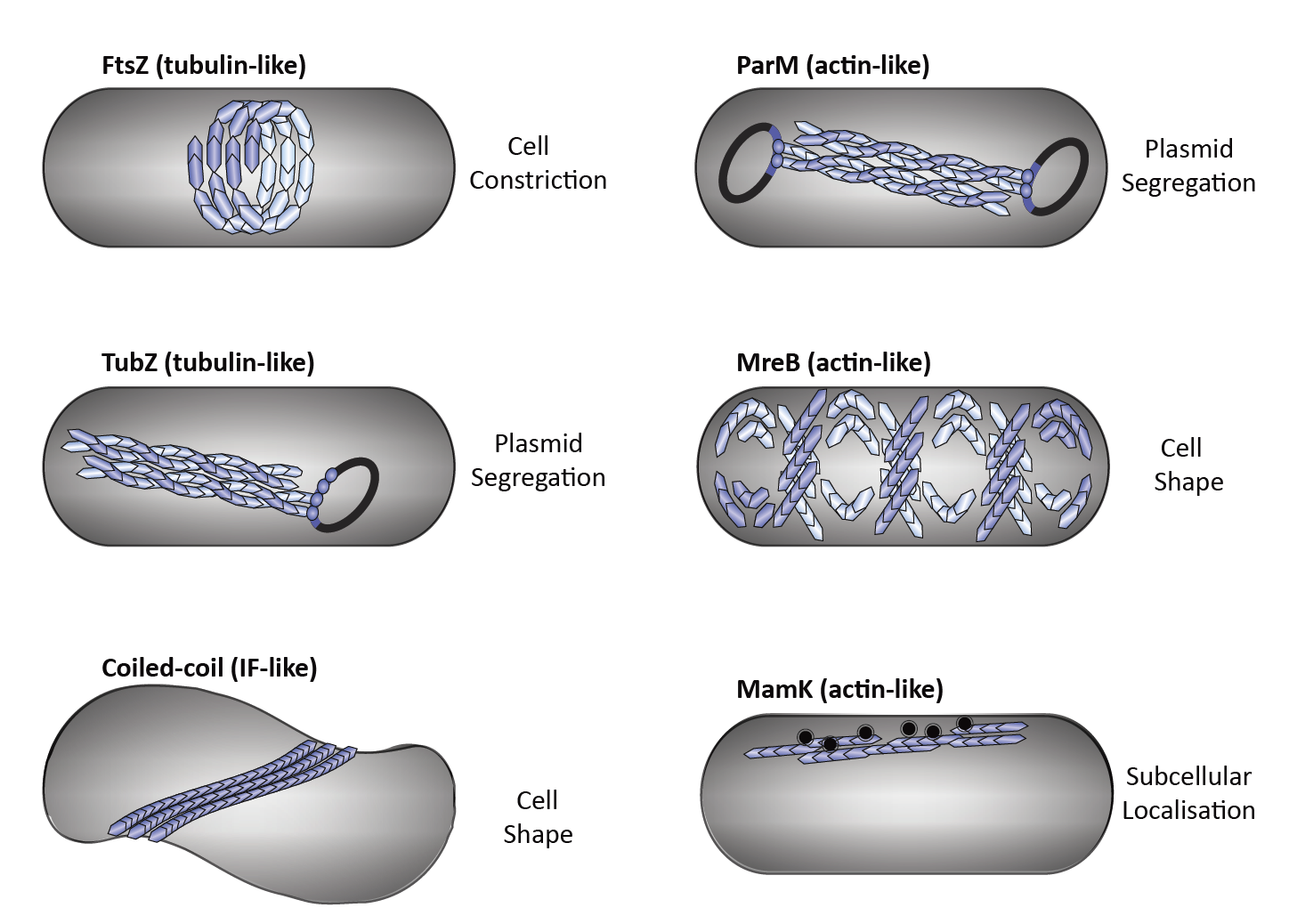
Fig. 3: Diverse functions of cytoskeletal elements in bacteria. Bacterial tubulin homologs: FtsZ performs cytokinesis (a), TubZ executes plasmid segregation (b). Intermediate filament-like coiled-coil proteins (e.g., Crescentin and Bactofilins) regulate cell shape (c). Bacterial actin homologs: ParM executes plasmid segregation (d), MreB maintains cell shape (e) and MamK organizes magnetosome chains (f).
Our recent work showed that the bacterial Min proteins (MinC and MinD) negatively regulate the FtsZ-ring (hence divisome) assembly by forming a new class of alternating copolymeric cytoskeletal filaments. These filaments are unique and unrelated to any of the conventional cytoskeletal elements, i.e. actin, tubulin or intermediate filaments and are remotely ‘related’ to eukaryotic septins.

Fig. 4: MinCD co-crystal structure suggests MinCD together form a new class of alternating copolymeric cytomotive filament (Ghosal et al, Nature Communications).
We used hybrid methods including, X-ray crystallography, cryo-EM, super-resolution imaging and biochemical reconstitution to demonstrate that the MinCD filaments are essential for proper cell division and confirmed their existence in vivo. We also put forward a mechanistic model for how MinCD filaments might regulate FtsZ-ring assembly in cells.